
Analysis group*: this describes the sequencing strategy used for the sequence being aligned, those strategies being 'low coverage', 'high coverage', 'exon targeted' and 'exome'. Population*: the Sample population 3 letter code, this code is defined in README.populations. Mapping algorithm: For ILLUMINA this is bwa, again older files may have bfast for solid or ssaha or smalt for 454. Sequencing platform: all our current release should be ILLUMINA but old alignment files will have SOLID or LS454. You may also see chromN files which represent mappings to just that chromosome. Region, this is generally either mapped or unmapped, the mapped files represents all the mapping to the reference genome, the unmapped file represents all the unmapped reads.  Sample name: this matches column 10 in the sequence.index file and represents the individual all the sequences belong to. The name can be broken down into 7 pieces: The bam filenames themselves contain a lot of information, e.g: The exome alignments also have a HsMetrics files which contain the results from Mapped basepairs in Gb is also shown as break down by population at the bottom of the file.Ģ0091216.alignment_stats.csv - a summary statistics of the very first release of main project BAMs. of individuals with > 10 Gb of mapped sequences. The file contains the following information break down by platform. Yyyymmdd_yyyymmdd.alignment_stats.csv - a summary statistics of BAMs in any two releases as specified by the dates in the file name a comparison of the two releases is captured in the "diff" values of the file. gz - a collective bas file of all BAM files. The following files about BAM statistics: With the latest date and identical to the alignment.index one level up), as wellĪs alignment.index files of previous BAM releases. There is an alignment.index and an which links the alignment bamįiles together with their matching bai file and bas file and gives md5sums for each.ĭirectory alignment_indices/ contains the most current alignment.index (the one Where the XXXXXXXX identifier is the sample name.
Sample name: this matches column 10 in the sequence.index file and represents the individual all the sequences belong to. The name can be broken down into 7 pieces: The bam filenames themselves contain a lot of information, e.g: The exome alignments also have a HsMetrics files which contain the results from Mapped basepairs in Gb is also shown as break down by population at the bottom of the file.Ģ0091216.alignment_stats.csv - a summary statistics of the very first release of main project BAMs. of individuals with > 10 Gb of mapped sequences. The file contains the following information break down by platform. Yyyymmdd_yyyymmdd.alignment_stats.csv - a summary statistics of BAMs in any two releases as specified by the dates in the file name a comparison of the two releases is captured in the "diff" values of the file. gz - a collective bas file of all BAM files. The following files about BAM statistics: With the latest date and identical to the alignment.index one level up), as wellĪs alignment.index files of previous BAM releases. There is an alignment.index and an which links the alignment bamįiles together with their matching bai file and bas file and gives md5sums for each.ĭirectory alignment_indices/ contains the most current alignment.index (the one Where the XXXXXXXX identifier is the sample name. 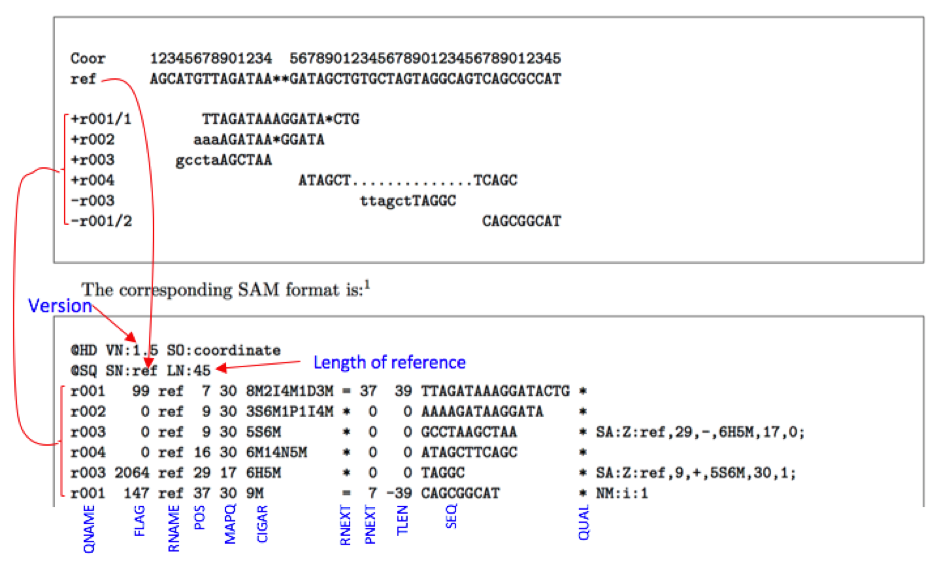
The alignments are found under data/XXXXXXXX/alignment and data/XXXXXXX/exome_alignment Of bas file is described further down in in section D. bai and a statistics file which has the same name but end with. Has two associated files: a index file which has the same name but ends with This is the binary version of the SAMįormat which is described here. Processed and what summary information is available for each alignment.Īll alignment data is in the BAM format.

This README describes the alignment data available on the ftp site, how it is







 0 kommentar(er)
0 kommentar(er)
